Modular Machine Learning and Classification Toolbox for ImageJ 3
Context
The Active segmentation ImageJ plugin was developed in the scope of GSOC 2016 and 2017. The plugin provides a general-purpose environment that allows biologists and other domain experts to use transparently state-of-the-art techniques in machine learning to improve their image segmentation results. ImageJ [1] is a public domain Java image processing program extensively used in life and material sciences. The program was designed with an open architecture that provides extensibility via plugins.
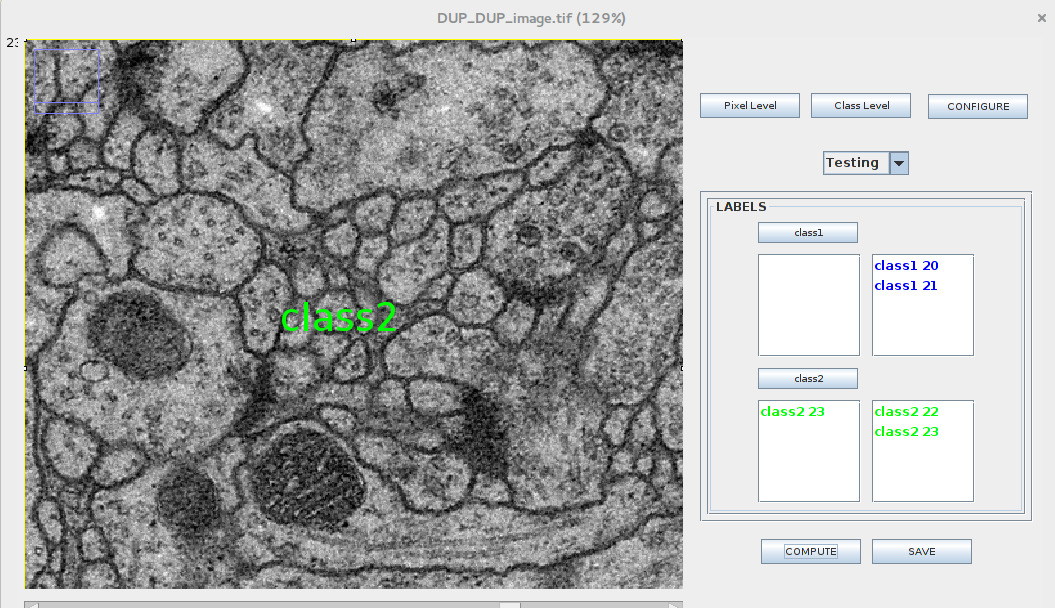
Motivation: We would like to expand the existing functionality of the Active Segmentation plugin to incorporate learning from entire images presented as instances. In GSOC 2017 we incorporated Zernike moments as a compound feature vector and used it to train the classifier.
Project idea: The present project can go into different directions: either to incorporate Legendre moments and parallelize their computation to further enrich the feature space or to incorporate Principal Component Analysis reduction scheme to reduce feature redundancy.
Tasks
- Fix existing issues and bugs
- Improve the user interface for region of interest display
- Extend the metadata format
- Provide a reference implementation
Minimal set of deliverables
- Requirement specification – Prepared by the candidate after understanding the functionality.
- System Design – Detailed plan for development of the plugin and test cases.
- Implementation and testing – Details of implementation and testing of the plugin.
The candidate
Required skills: Experience with Java
Desired skills: experience with ImageJ, machine learning preferably WEKA
Mentors: Dimiter Prodanov ([email protected]), INCF Belgian Node; (backup) Sumit Vohra, KU Leuven
